
The New Code of Life: How GPT Architectures Are Revolutionizing Biological Research
For years, the discourse surrounding Large Language Models (LLMs) has been dominated by their remarkable ability to understand and generate human language. From crafting emails to writing code, models like GPT-4 have reshaped our digital interactions. However, the latest wave of GPT Research News reveals a far more profound application: deciphering and rewriting the very language of life itself. The same transformer architecture that powers ChatGPT is now being applied to the complex sequences of proteins and DNA, unlocking unprecedented capabilities in drug discovery, synthetic biology, and personalized medicine. This convergence of artificial intelligence and life sciences marks a pivotal moment, moving beyond text generation to biological generation.
This emerging field represents a paradigm shift. Instead of treating biology as a discipline of observation and experimentation alone, we can now approach it as a generative design problem. The latest OpenAI GPT News and developments from other leading research labs suggest a future where scientists can prompt an AI to design a novel enzyme or therapeutic protein with specific functional properties. This article delves into the technical underpinnings of this revolution, exploring how GPT models are being adapted for biological data, the groundbreaking applications this enables, and the critical challenges and ethical considerations that lie ahead in this new frontier of computational biology.
The Transformer’s New Language: From Human Text to Biological Code
The application of GPT-like models to biology is not merely a clever analogy; it’s rooted in a fundamental similarity between language and biological sequences. This conceptual leap is driving some of the most exciting GPT Trends News in the scientific community, promising to accelerate research in ways previously unimaginable.
Why LLMs for Biology? The Sequence-as-Language Paradigm
At its core, a protein is a long chain of amino acids, akin to a sentence made of letters. The specific sequence of these amino acids dictates how the protein folds into a complex three-dimensional shape, and this shape, in turn, determines its function. For decades, predicting this structure and function from a sequence has been a central challenge in biology. The transformer architecture, the foundation of all modern GPT models, is uniquely suited to tackle this problem. Its key innovation, the self-attention mechanism, allows the model to weigh the importance of all other elements in a sequence when processing a single element. In language, this means understanding that the word “it” in “The robot picked up the ball because it was heavy” refers to the ball, not the robot. In biology, this translates to understanding that two distant amino acids in a sequence might be critically close in the final folded structure, forming an active site. This ability to capture long-range dependencies is what makes the latest GPT Architecture News so relevant to proteomics and genomics.
Key Innovations in Generative Biology
While models like AlphaFold have revolutionized protein structure prediction, the new wave of generative models aims to go a step further. Instead of just predicting the structure of a known sequence, these bio-LLMs can generate entirely novel sequences with desired properties. Recent research showcases specialized models, sometimes referred to as “micro” versions of larger architectures like GPT-4, that have been trained specifically on massive biological datasets. The process involves extensive GPT Fine-Tuning News, where a base model is adapted using curated datasets from protein databases like UniProt. The goal is to create GPT Custom Models News that can respond to functional prompts, such as “Generate a thermostable enzyme that catalyzes reaction X.” This moves the field from analysis to synthesis, providing researchers with a powerful new tool for engineering biology from the ground up. This generative capability is a central theme in recent GPT-4 News and a likely focus for future GPT-5 News.
Under the Hood: How GPT Models Learn the Language of Life
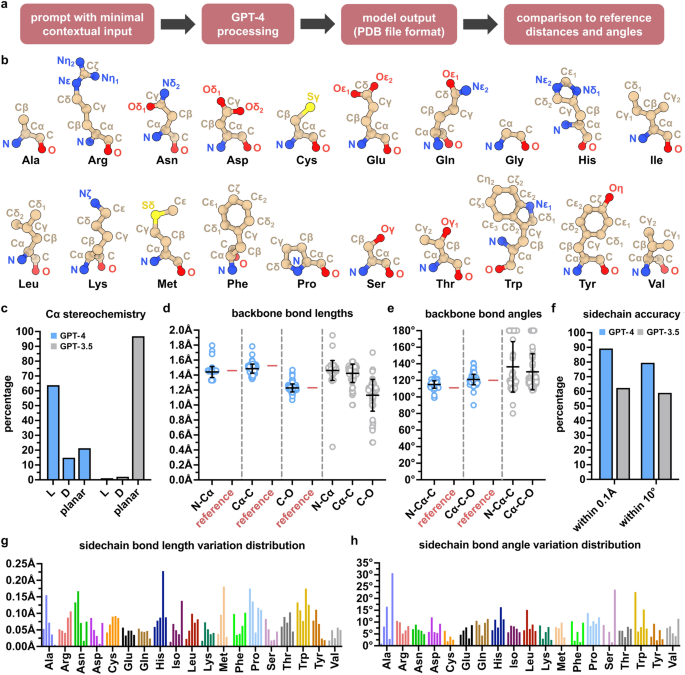
Adapting a model designed for human language to the intricate world of molecular biology requires significant technical modifications to its architecture, tokenization, and training process. Understanding these details is key to appreciating both the power and the limitations of these emerging technologies.
Adapting the Transformer Architecture for Biological Data
The core transformer block remains the same, but the input and output layers are re-engineered for biological sequences. The first critical step is tokenization. In natural language processing, text is broken down into words or sub-words. For proteins, the most straightforward approach is to assign a unique token to each of the 20 common amino acids. This is a hot topic in GPT Tokenization News, as more advanced strategies might involve tokenizing common amino acid motifs or “words” to capture more biological context. The model must also understand the linear order of the sequence, which is handled by positional encodings. A significant area of research covered in GPT Scaling News involves finding the right model size. While a massive model offers greater capacity, it can be computationally expensive for inference. This has led to a focus on GPT Efficiency News, with techniques like GPT Compression News and GPT Quantization News being explored to create smaller, faster, yet powerful models suitable for widespread deployment in research labs.
The Crucial Role of Training Data and Fine-Tuning
The performance of any AI model is contingent on the quality and scale of its training data. For bio-LLMs, this means leveraging decades of accumulated biological knowledge stored in public databases. The pre-training phase typically involves feeding the model millions of known protein sequences, allowing it to learn the fundamental “grammar” of protein structure and function without explicit labels. This is a core concept in GPT Training Techniques News. Following this, the model is fine-tuned on more specific tasks. For example, to create a model that designs antibodies, it would be fine-tuned on a dataset of known antibody sequences and their binding targets. This highlights the importance of high-quality, well-annotated GPT Datasets News for developing specialized and effective models. The ability to fine-tune these models is what transforms them from general sequence predictors into powerful tools for specific scientific challenges, a key theme in GPT Applications News.
The Ripple Effect: Applications and Implications for Science and Medicine
The theoretical potential of generative biological models is rapidly translating into tangible, real-world applications. From the pharmaceutical industry to materials science, these AI systems are poised to dramatically accelerate the pace of innovation, bringing us closer to solving some of humanity’s most pressing challenges.
Revolutionizing Drug Discovery and Therapeutics
The most immediate and impactful application lies within the realm of healthcare, a constant source of GPT in Healthcare News. Consider the development of a new therapeutic drug. Traditionally, this is a multi-year, billion-dollar process of screening millions of compounds. With a generative model, a researcher could bypass much of this.
Real-World Scenario: A team is developing a treatment for a new viral pathogen. They need an antibody that can neutralize the virus by binding to its spike protein. Instead of laborious lab-based screening, they could use a specialized bio-LLM. Their prompt might be: “Generate 1,000 novel, high-affinity human antibody variable region sequences that target epitope XYZ on the viral spike protein, optimized for stability and low immunogenicity.” The model would output a list of promising DNA sequences. These sequences could then be synthesized and tested in the lab, reducing the discovery phase from years to weeks. This streamlined workflow, potentially accessed via secure GPT APIs News, represents a seismic shift in pharmaceutical R&D and is a major focus of current GPT Research News.
Beyond Pharmaceuticals: Biomanufacturing and Materials Science
The applications extend far beyond medicine. In industrial biotechnology, scientists are using these models to design novel enzymes for a variety of purposes. This could include creating enzymes that can efficiently break down plastics to combat pollution, or developing more effective biofuels by engineering enzymes that convert biomass into ethanol. This falls under the broader umbrella of GPT Applications in Content Creation News, where the “content” is a functional molecule. In materials science, researchers could prompt for proteins that self-assemble into novel materials with unique properties, like ultra-strong, lightweight fibers or biocompatible scaffolds for tissue engineering. These advancements showcase the versatility of the underlying GPT architecture.
Navigating the Ethical Minefield
With great power comes great responsibility. The ability to generate novel, functional proteins raises significant ethical and safety concerns. This has become a critical topic in GPT Ethics News and GPT Safety News. A malicious actor could theoretically attempt to design harmful pathogens or toxins. This necessitates a robust framework for GPT Regulation News, including strict access controls for the most powerful models and “know your customer” protocols for services that synthesize DNA from digital sequences. Furthermore, issues of GPT Bias & Fairness News are relevant; if training data over-represents biology from certain populations, the resulting therapeutics might be less effective for others. Ensuring data privacy, as discussed in GPT Privacy News, is also paramount when dealing with sensitive genetic information.
Navigating the New Frontier: Challenges and Strategic Recommendations
While the promise of generative biology is immense, the path forward is not without significant technical hurdles and practical challenges. Researchers and organizations looking to leverage these technologies must adopt a strategic approach grounded in best practices and a clear understanding of the current limitations.
Overcoming the “Hallucination” and Validation Problem

A well-known pitfall of LLMs is their tendency to “hallucinate”—that is, to generate confident but factually incorrect or nonsensical output. In biology, a hallucination translates to a protein sequence that is biologically non-viable; it may not fold into a stable structure or perform the intended function. This is a major challenge.
Best Practice: Treat AI-generated sequences as highly educated hypotheses, not proven solutions. The output of a bio-LLM is the starting point of the scientific method, not the end. A critical best practice is to establish a tight feedback loop between computational generation and empirical validation. This involves:
- Generating a diverse set of candidate sequences with the model.
- Using computational filters and structure prediction tools (like AlphaFold) to screen for the most promising candidates.
- Synthesizing the top candidates and testing their properties in the lab (in vitro).
- Using the experimental results to further fine-tune the generative model, improving its accuracy over time.
This iterative cycle is essential for success and is a key area of development in GPT Benchmark News and GPT Inference News.
The Imperative for Multimodality
The future of AI in biology is multimodal. A protein’s function is not determined by its amino acid sequence alone. It is a complex interplay of sequence, 3D structure, cellular location, and interaction partners. The next generation of models, a topic of frequent speculation in GPT Multimodal News, will need to integrate these different data types. Imagine a model that can process a protein sequence, a 3D structural file (linking to GPT Vision News), and a description of its functional context simultaneously. Such a model could generate far more sophisticated and reliable outputs. This convergence of data is a central theme in GPT Future News and will require advances in everything from GPT Hardware News to the underlying model architectures.
Conclusion
The latest GPT Research News signals a profound expansion of artificial intelligence from the digital realm of language and code into the physical realm of biology. The application of transformer architectures to protein and gene sequences is not an incremental improvement; it is a transformative leap that redefines the boundaries of what is possible in life sciences. By treating biological sequences as a language, we can now move from simply reading the book of life to actively writing new chapters. The potential to accelerate drug discovery, design sustainable biomaterials, and develop personalized therapies is immense, promising to reshape industries and improve human health on a global scale.
However, this journey is just beginning. Realizing this potential will require overcoming significant technical challenges, navigating complex ethical landscapes, and fostering deep collaboration between AI researchers and domain experts in biology. The convergence of these fields is creating a vibrant GPT Ecosystem News, where innovation is driven by a shared vision of a healthier, more sustainable future engineered, in part, by the very models that are reshaping our world.



